| dc.contributor.author | Jordan, Bertrand | - |
| dc.date.accessioned | 2025-10-28T17:13:02Z | |
| dc.date.available | 2025-10-28T17:13:02Z | |
| dc.date.issued | 2024 | |
| dc.identifier.citation | Jordan, Bertrand ; L’édition de nouvelle génération, Med Sci (Paris), Vol. 40, N° 11 ; p. 869-871 ; DOI : 10.1051/medsci/2024149 | |
| dc.identifier.issn | 1958-5381 | |
| dc.identifier.uri | http://hdl.handle.net/10608/15225 | |
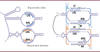
| dc.description.abstract | Recent work on bacterial insertion sequences reveals that some of them use an RNA sequence (called Bridge RNA or Seek RNA) to define both donor and target DNA specificity. This opens the way to easy insertion of kilobase DNA sequences at pre-defined sites in the genome, announcing a host of new possibilities. The system still needs a lot of tweaking, as it has only been demonstrated in bacteria, but it holds great promise for genome editing and engineering. | en |
| dc.language.iso | fr | |
| dc.publisher | EDP Sciences | |
| dc.relation.ispartof | Forum | |
| dc.rights | Article en libre accès - License CC BY 4.0 | fr |
| dc.rights | Médecine/Sciences - Inserm - SRMS | fr |
| dc.rights.uri | http://creativecommons.org/licenses/by/4.0 | |
| dc.source | M/S. Médecine sciences [ISSN papier : 0767-0974 ; ISSN numérique : 1958-5381], Vol. 40, N° 11; p. 869-871 | |
| dc.subject.mesh | Animaux | fr |
| dc.subject.mesh | Humains | fr |
| dc.subject.mesh | Bactéries | fr |
| dc.subject.mesh | génétique | fr |
| dc.subject.mesh | Systèmes CRISPR-Cas | fr |
| dc.subject.mesh | Édition de gène | fr |
| dc.subject.mesh | méthodes | fr |
| dc.title | L’édition de nouvelle génération | fr |
| dc.title.alternative | New-generation editing | en |
| dc.type | Article | |
| dc.contributor.affiliation | Biologiste, généticien et immunologiste, Président d’Aprogène (Association pour la promotion de la Génomique) , 13007 Marseille , France | |
| dc.identifier.doi | 10.1051/medsci/2024149 | |
| dc.identifier.pmid | 39656986 | |